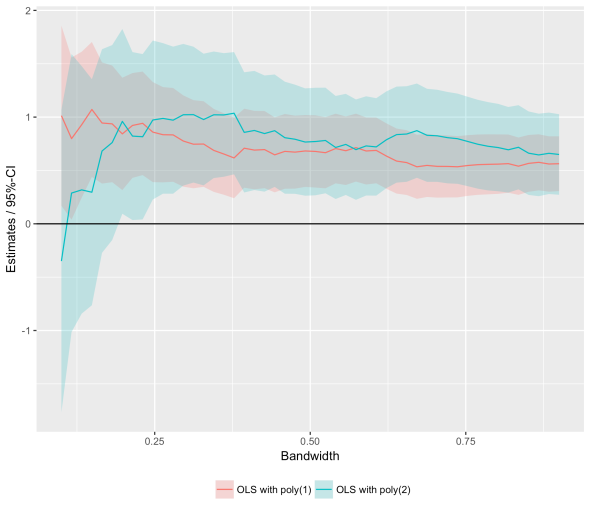
Bandwidth Plot with purrr + ggplot2
Posted: February 14, 2017 Filed under: Post | Tags: R Leave a commentRDD estimates are sensitive to bandwidth choices which is why one typically plots them for different bandwidths. Here is some code to quickly obtain RDD estimates for different bandwidths and plot the results. The code relies on the amazing purrr package.
library(purrr)
library(ggplot2)
library(dplyr)
library(broom)
library(stringr)
# Define a helper function that gives back local linear
# regression estimates when handed a bandwidth and a specification
get_lm_bw_est <- function(df,bw,formula) {
return( df %>% filter( abs(runningvar) <= bw ) %>%
do(tidy(lm(formula, data=.))) %>%
filter(term=="treat") %>%
mutate(bw=bw, spec=format(formula) ) )
}
set.seed(42)
# Sim Data
x <- runif(1000,-1,1)
y <- 5+ 3*x + 0.5*(x>=0) + rnorm(1000)
fakedata <- data.frame(y=y,runningvar=x, treat=as.numeric(x>=0) )
# Set up grid of specification choices and bandwidths
spec1 <- y ~ treat * runningvar
spec2 <- y ~ treat * (runningvar + I(runningvar^2))
spec <- c(spec1,spec2)
simframe <- expand.grid(bw=seq(.1,.9,length=50), spec=spec)
# Estimate, i.e. map simframe values to helper function
m <- with(simframe, map2_df(bw, spec, ~ get_lm_bw_est(fakedata, .x, .y) ))
# Calculate 95% CI and plot results
m <- m %>% mutate(lo=qnorm(0.975)*std.error + estimate,
hi=qnorm(0.025)*std.error + estimate)
m <- m %>% mutate(spec=str_count(spec, "running"),
spec=paste("OLS with poly(", spec, ")", sep=""))
ggplot(m, aes(bw,estimate,group=spec)) +
geom_ribbon(aes(ymin=lo,ymax=hi, fill=spec), alpha=0.25) +
geom_line(aes(colour=spec)) + geom_hline(aes(yintercept=0)) +
theme(legend.position='bottom', legend.title=element_blank()) +
ylab("Estimates / 95%-CI") + xlab("Bandwidth")
Zero-Numerator Problem: Calculating the Expected Number of Mistakes in Data Entry Jobs
Posted: January 1, 2017 Filed under: Post | Tags: Bayesian, data entry, R, research practice Leave a commentI have a project for which I need to digitize a series of tables from scanned pdf pages. Due to the scan quality, some pages are handled manually by research assistances. After receiving a new badge, I typically like to estimate how many data-entry errors I have to expect. Therefore I draw a random evaluation sample and carefully check if the entered numbers matches the originals in the pdf. Depending on the sample size and the quality of the badge, I often arrive at a count of zero errors for the evaluation sample.
What does a zero count for the evaluation sample say about the distribution of errors in the entire badge? If the sample is randomly drawn, the number of errors, F, has a Bernoulli density with parameter S and p. The parameter S denotes the number of checked cells (the sample size) and p measures the true proportion of incorrect values across all cells. While S is known, p is not and constitutes the estimand.
A typical estimator for p is the sample mean, but in the event of F=0, the estimate for p is 0 for any sample size. That is, the sample mean estimator always suggests that the expected number of errors is zero — independent of the size of the evaluation sample. The same holds for an interval estimator of the 95% confidence interval. In the event of zero errors, the estimate for the sample variance is also ‘0’ and consequently the 95% confidence interval is zero regardless of the sample size.
A version of this problem is well-known in the bio-statistical literature. The task here is to compare two procedures (or treatments): One for which the risk of people being harmed (killed) is well-known and another for which only sample information with not a single harmed individual is available. The standard approach seems to be to calculate the upper bound of the 95% confidence interval using 3/S as an estimator (also known as The Rule of Three).
As pointed out in several contributions — for a summary see Winkler et al. (2002)
— a more principled way to form an estimate requires Bayesian Statistics and starts with the assignment of an informed prior density over p. This prior is then updated using the newly gathered data point ‘0 errors in an evaluation sample of size S’. To the extend that the prior is expressed as a Beta distribution, the calculations are algebraically easy.
However, the tricky part is typically to come up with a reasonable prior density for p. In practice, I make use of some very old numbers reported in a paper by Baddeley and Longman (1978). Back in the days when post codes had to be manually typed, they conducted an experiment with groups of postmen to evaluate what type of training procedures reduce the number of wrongly typed “alpha-numeric code material”.
In the table below I reproduce the Baddeley-Longman numbers and also the variance I calculated from the range estimates. Averaging across the four groups suggests that the rate of errors is about 1.4% with a variance 0.7. A more conservative set of estimates might be stipulated from using the values reported from group 4 alone (2.1 for the mean value and 1.1 for the variance).
| Group 1 | Group 2 | Group 3 | Group 4 | |
|---|---|---|---|---|
| % Incorrect | 1.09 | 1.14 | 1.41 | 2.06 |
| Range | 0.22 – 2.18 | 0.06 – 2.42 | 0.40 – 3.45 | 0.38 – 4.65 |
| N | 18 | 18 | 18 | 18 |
| Variance | 0.49 | 0.59 | 0.76 | 1.07 |
Using these values, it is easy to calculate the implied Beta prior and the posterior distribution and its characteristics when combined with the information from the evaluation sample. In Bayesian Statistics the calculations are known as Beta-Binomial update.
In order to simplify these calculations, I have written a short R function that makes the calculations when given information about the evaluation sample. The user can choose between the two pre-programmed priors mentioned above or supply its own. The function is part of my datatools package and can be installed from my github, via devtools::install_github('sumtxt/datatools').
As an example, suppose we have an evaluation sample of S=20 with F=0 (no errors). Using the function, we find that the expected proportion of errors in the population is about 1.3% and the upper bound of the 95% credible interval equals 2,5%.
datatools::est_typing_err(20, F=0, prior='postmen', quantity='mean') [1] 0.01306888 datatools::est_typing_err(20, F=0, prior='postmen', quantity='95q') [1] 0.02538156
Notice: These estimates differ from the estimates implied by the sample-mean estimator (0%) and the rule-of-three estimator (3/20=15%).
IR dyad-year dataset with 10 lines of dplyr + tidyr code
Posted: July 29, 2015 Filed under: Post | Tags: dplyr, International Relations, IR, R, tidyr 2 CommentsDirected (country)-dyad-year datasets are quite common in International Relations (IR) research and from time to time one requires an empty one. Creating this used to be a hassle with +50 lines of code (see example in PERL or R). Luckily, with tidyr + dplyr + pipping building such a dataset requires at most 10 lines of code:
system <- read.csv("./raw data/states2011.csv")
system <- system %>% select(ccode, styear, endyear)
system <- system %>% expand(statea=ccode, stateb=ccode, year=seq(1816,2011)) %>%
filter(statea!=stateb) %>%
left_join(., system, by=c("statea"="ccode")) %>%
filter(year >= styear & year <= endyear) %>%
select(-styear,-endyear) %>%
left_join(., system, by=c("stateb"="ccode")) %>%
filter(year >= styear & year <= endyear) %>%
select(-styear,-endyear)
First, the code block creates all possible country-country-year pairings (line 3) and then filters out the dyad-years which are inadmissible either because a) the dyad involves the same country (line 4) or b) at least one of the dyad-members does not exist in a particular year (lines 5-10).
Data Source: Correlates of War System Membership 2011
Tricks for working with JAGS
Posted: May 8, 2015 Filed under: Post | Tags: BUGS, JAGS, R Leave a comment- The Gamma distribution is frequently used as a prior for the precision in a model because of its conjugacy with the Normal. Given JAGS parameterization of the Gamma distribution with a rate and shape parameter, it’s not easy to select sensible hyper-parameters. It’s easier to think of hyper-parameters that correspond to the mean and the variance of a prior for the precision. To use mean and variance hyper-parameters (mu and sigma) with JAGS’s
dgamma, use:dgamma( ((mu^2)/sigma), (mu/sigma)). - The
glmmodule provides a series of more efficient samplers that JAGS automatically selects whenever possible (e.g. Albert/Chib (1993)). This usually leads to much better mixing. I tend to load the module right away with the actualrjagspackage, e.i.library(rjags); load.module("glm"). - To check which samplers are selected by JAGS, use the
list.samplers()function on the JAGS model object. This is useful, because sometimes a slight change in the coding helps JAGS to select better samplers. For example, when using a logit model in JAGS and you want the Holmes-Held sampler from the glm module to work, do not usedmnorm()butdnorm()for the prior of the coefficient, e.i. do not use blocking. - Avoid deterministic nodes as much as possible to increase speed. In particular, do all pre- and post processing in R including drawing from the predictive distribution and compute deterministic functions inside probabilistic statements, e.g.
y ~ dnorm( (x*beta), 1)instead ofmu . - For comprehensible / readable code, let the running index be the small letter corresponding to the capital letter of the upper limit, e.i.
for(n in 1:N)instead offor(i in 1:N). This becomes especially useful when one has many nested hierarchies.
Gibbs Samplers in R and Notes
Posted: May 4, 2015 Filed under: Post | Tags: MCMC, R Leave a commentI uploaded several R files and notes for Gibbs samplers to my Website’s resource page. They include: Factor Analysis, Finite Mixture of Linear Regression Models and Probit Regression. I have written all this a while ago when I was reading about these models. Most of the samplers are fairly standard and well-implemented efficiently in R packages calling c++ in the background. These packages are also referenced in the R files. Nevertheless, the R code might be useful for pedagogical purposes or as building block for more complex schemes.
Generate a panel version of the UCDP/PRIO Armed Conflict dataset
Posted: October 30, 2013 Filed under: Post | Tags: data, R, UCDP 1 CommentI spent the morning writing a short R script that transforms the UCDP/PRIO Armed Conflict dataset into a panel dataset in which each conflict is observed from the year it started to the year it ended (or the last year in the dataset). The unit of observation in the original dataset is an active conflict observed in a year t. More precisely, it is an ongoing conflict episode (for a particular conflict) in a year t. Inactive years between conflict episodes are not recorded (see figure with an example from Ethiopia and two variables from the dataset). The purpose of the code below (especially the second part) is to fill-in the observations between conflict episodes. In addition to the panel structure, I wanted to have a more natural variable structure / variable names (this is what the first part is doing).
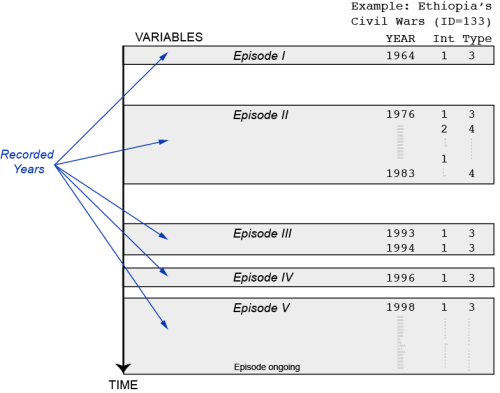
The code below might not be optimal for all applications, but it is probably still useful for others that need to get a panel version of the UCDP/PRIO. If you spot mistakes or have ideas for improvements, please share them in the comments. For an introduction to the dataset, definition of variables etc, please read the original codebook.
Read the rest of this entry »
Drawing games with TikZ
Posted: July 14, 2013 Filed under: Post | Tags: game, Latex, tikz Leave a commentTikZ is a great TeX package to draw whatever you feel like drawing. I just used it to draw game trees for a paper. A good starting point for doing this was a tutorial by Haiyun K. Chen from Simon Fraser University. Below is the code I came up with for my games. The first game is an extensive form game among two players that have to choose twice simultaneously between two alternatives. The second game is also an extensive form game, but among three players. In the first stage, player C has to choose a value for z. Next, the two players E,F have to choose among two alternatives simultaneously.
Gnuplot and the Beta Distribution
Posted: June 25, 2013 Filed under: Post | Tags: distribution, stats, visualization Leave a commentI recently discovered gnuplot, a command-line plotting tool. I discovered it while trying to get some work done with Maxima, a Computer Algebra System Software. Gnuplot doesn’t make plots as pretty as R’s ggplots or d3 – but it is flexible and you get visual impressions of your math functions quickly. Here is a little animation that I have done with the Beta distribution: code and result.
Running Parallel MCMC without specific R packages
Posted: May 19, 2013 Filed under: Post | Tags: MCMC, R 1 CommentRunning multiple, distinct Markov chains that have a posterior density as their equilibrium density is useful to use convergence diagnostics such as the Gelman / Rubin diagnostic (Gelman & Rubin, 1992). In order to save time, one likes to run each Markov chain in parallel. There are a number of packages in R (parallel, snow,…) to run code in parallel. However, I find it more useful to write a script for a single chain and then dispatch it multiple times from the terminal. Here is an example with the MCMCpack:
Save the following as “sim.R”:
substream <- as.numeric(commandArgs(trailingOnly = TRUE))
library(MCMCpack)
data(birthwt)
filename <- paste("posterior_", substream, ".Rdata" , sep="", collapse="")
out <- MCMCprobit(low~as.factor(race)+smoke, data=birthwt,
b0 = 0, B0 = 10, seed=list(NA, substream))
save(out,file=filename)
The first line grabs the seed argument from the terminal and pass it to MCMCprobit function which generates a single chain. The script can be executed from the terminal with four different substream seeds as follows:
Rscript --arch=x86_64 sim.R 1 & Rscript --arch=x86_64 sim.R 2 & Rscript --arch=x86_64 sim.R 3 & Rscript --arch=x86_64 sim.R 4
If you don’t have a 64-Bit machine, use “x86_32”. Provided that you have four cores, one core will be used for each of these jobs. Next, you can fuse the output together in an mcmc.list object and run the Gelman/Rubin diagnostic.
library(coda)
load("posterior_1.Rdata")
out1 <- out
load("posterior_2.Rdata")
out2 <- out
load("posterior_3.Rdata")
out3 <- out
load("posterior_4.Rdata")
out4 <- out
posteriors <- as.mcmc.list(list( as.mcmc(out1), as.mcmc(out2),
as.mcmc(out3), as.mcmc(out4)))
gelman.diag(posteriors)
The great benefit of this approach is that the R code is less crammed with parallelization functions and the process can be monitored since the output is dumped right into the terminal window.
References
Gelman, Andrew & Donald B. Rubin. 1992. “Inference from Iterative Simulation Using Multiple Sequences.” Statistical Science 7(4):457–472.
Political Science Job Market – We need a model!
Posted: April 28, 2013 Filed under: Post | Tags: model, political science Leave a commentAPSA recently published some ugly plots on the number of Job Postings in APSA e‐Jobs. I wonder why we don’t have a predictive model for that? It should be possible to predict the number of Job postings based on some indicators (US GDP growth, NSF budget, etc.). Anyone?